Stephen A. Lauer
Biostatistician
Greetings!
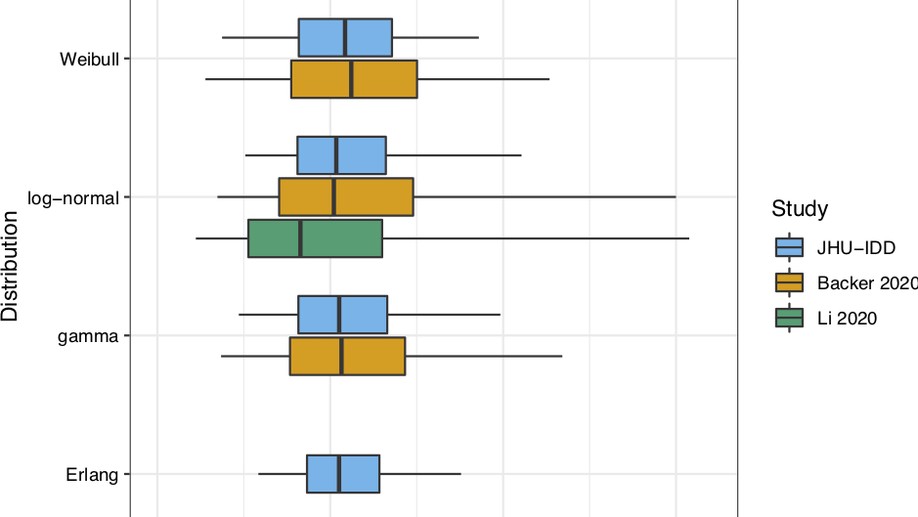
I am a postdoctoral researcher in the Infectious Disease Dynamics Group at Johns Hopkins Bloomberg School of Public Health. My current research focuses on estimating the burden and distribution of cholera in Bangladesh based on serological biomarkers. As a side project, I have estimated some of the epidemiological parameters of the recent 2019-nCoV outbreak.
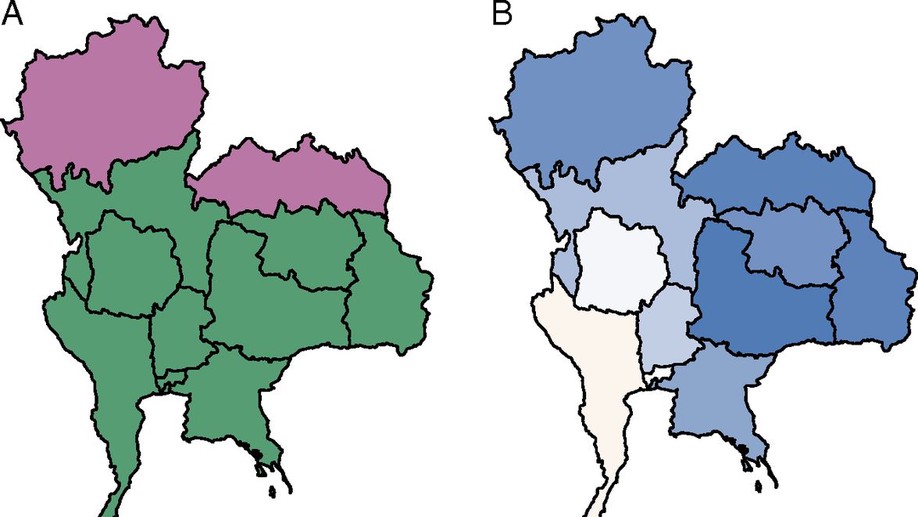
During my PhD at UMass, I worked with the Thailand Ministry of Public Health to forecast dengue fever incidence at short and long time scales. During my time as a grad student, I developed a number of Shiny web applications for various projects (SEIGMA, dengue predictions, and ALERT).
Outside of the office, my interests include sports, travel, economics, voting reform, transportation issues, and video games. Check out my full bio for more details!
Specialties
- Probabilistic Forecasting
- Causal Inference
- Data Analysis and Visualization
Education
-
PhD in Biostatistics, 2019
University of Massachusetts, Amherst
-
MS in Biostatistics, 2014
University of Massachusetts, Amherst
-
BS in Business, Operations Management, 2009
University of Maryland, College Park